EECS545 Coursework Project
Duration: 02/2022-05/2022
Teammates: Haowen Chen, Yuang Lu, Yuqi Yan, Ying Yuan
Introduction
During Diagnosis, treatment and surgery with computer aid, medical image segmentation plays a crucial part in identifying pixels of anatomical objects in medical images. However, it is a time-consuming and heavy work to conduct segmentation on images one by one by doctors. The primary challenges for segmentation mainly come from three aspects: (1) Complex boundary interactions, (2) Large appearance variation, (3) Low tissue contrast. These challenges. With the advance of deep learning, it is necessary to conduct new techniques to help medical image segmentation. [1]
In this project, we focused on the abdominal multi-organs segmentation problem. We proposed two novel neural network structures to solve two main problems: fragmental objects edges and failure to detect small, direction-dependent objects. The performance of medical image segmentation is expected to be enhanced by fusing Self Attention mechanism and convolutional neural network.
Related Work
In this section, recent work for medical image segmentation will be gone through. To achieve pixel-wise classification, researchers proposed the encoder-decoder network structure. UNet is one of the most outstanding works among them. Based on the structure, novel techniques have been introduced into UNet. [2]
ResUNet
Residual UNet(ResUNet) [3] is inspired by ResNet. [4] Previous results showed that with the number of network layers increased, the feature identities and performance will be lost. It is caused by vanishing gradients in deep networks. By concatenating features, the skip connection before downsampling or upsampling in ResUNet helps preserve feature maps.
According to our results, ResUNet is capable of detecting all 13 organs in the Synapse dataset including small and directional objects such as the left and right adrenal gland. However, all other transformer-based models failed to do so. It can also perform better in segmenting other small organs such as portal vein and splenic vein. It is believed that the convolutional layer helps the model to detect and segmentate these small objects.
TransUNet
TranUNet is a model based on Transformers and UNet, leveraging both detailed high-resolution spatial information from CNN features and the global context encoded by Transformers. Transformer serves as a strong encoder, providing an attentive feature sequence. According to former results, this architecture achieves superior performance over any other method on medical image segmentation. Therefore, TransUNet works as the baseline model in this project and implements a novel model. [5]
Swin-UNet
Based on the outstanding performance in Trans-UNet, optimization of Transformer might be opt for image segmentation. Swin-UNet could be one among them. Different from all silices connected together in Trans-UNet, the limited slices are connected with the neighboring slices in Swin-UNet. When in the higher layer, the output from the neighbor transformer will be the input of the next layer. By this way, more details from the images could be trained and neighboring information could affect each other. [6]
Though Swin-UNet has higher DICE score over TransUNet overall, which means that it could better classify each pixel in the right class, the results showed that the segmented objects trained by Swin-UNet are fragmental and have rougher edges compared to TransUNet.
UTNet
Although transformer variants perform well in vision tasks, pixels packed in the same input image patch do not contain enough spatial information, leading to weaker inductive bias compared with traditional CNN. Therefore, vision transformer-based models require a large image set to gain enough prior knowledge in the image domain. However, a major characteristic of medical image segmentation tasks is that they usually cannot provide such a big dataset, using pretrained weight on other large image datasets then becomes a widely adopted solution.
UTNet, unlike other models, employs another method. Common vision transformers and their variants pack pixels into patches and flattening them into vectors while UTNet preserves the first few encoding layers as convolutional layers, the pixel-level spatial relations thus can be retained. [7]
UNETR
UNETR proposed a novel transformer-based model for volumetric medical image segmentation. Instead of extracting features with Transformers at the bottleneck of U-Net, UNETR has its skip-connected decoder that combines representation from multiple intermediate Transformers layers. Deconvolutional layers are applied to these intermediate representations to increase the resolution. Such architecture helps the network capture global contextual representation at multiple scales and increase the capability for learning long-range dependencies. [8]
TransFuse
In order to improve the efficiency of modeling global contexts and preserve low-level features, TransFuse uses a parallel approach to merge Transformers and CNNs. This allows the efficient collection of both global dependencies and low-level spatial features in a significantly shallower manner. The BiFusion module is a novel fusion technique that efficiently fuses the multi-level features from both branches. According to the research, TransFuse obtains state-of-the-art results on both 2D and 3D medical picture sets with significantly fewer parameters and significantly faster inference. [9]
Proposed Method
In our novel models, Swin-Transformer and UTNets act as encodes to learn sequence representations of the input and further enhance the efficiency in capturing the global information. The overall diagram of the proposed model is shown in Fig. 1.
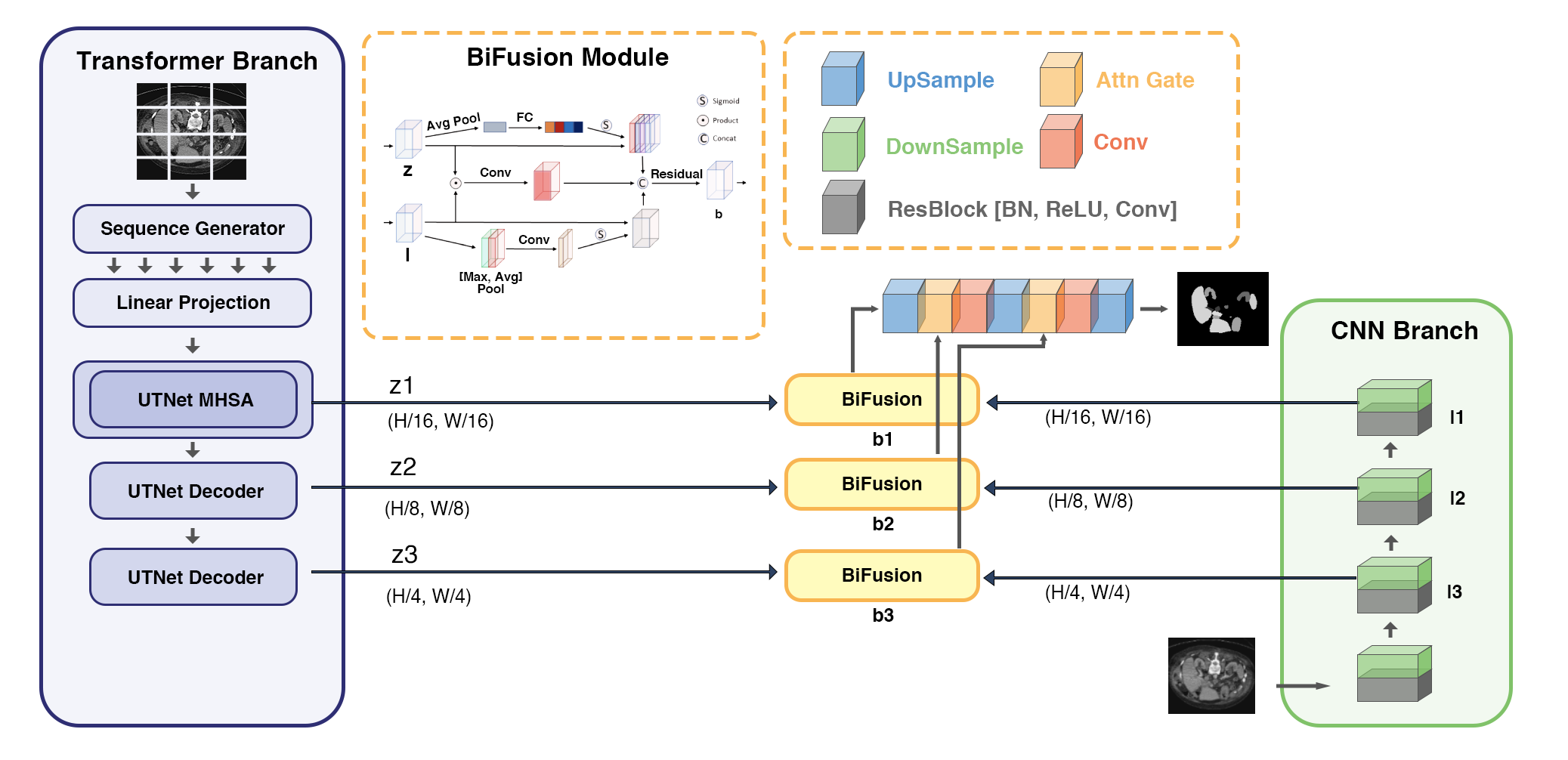
Fig 1. - Pipeline
This model consists of two parallel branches that process information differently
1. CNN Branch
It gradually increases the receptive field and encodes the feature from local to global. With the Transformer branch to obtain global context information instead, CNN branch could be shallower, and the proposed model could be also retaining richer local information. The output of the 3rd block ($\textbf{l}^1\in \mathbb{R}^{\frac{H}{16} \times \frac{W}{16}\times C_1 }$), 2nd block ($\textbf{l}^2\in \mathbb{R}^{\frac{H}{8} \times \frac{W}{8}\times C_2 }$), and 1st block ($\textbf{l}^3\in \mathbb{R}^{\frac{H}{4} \times \frac{W}{4}\times C_3 }$)) are fused with the results from Transformer. We propose different models with CNN variants.
2. Transformer Branch
It starts with global self-attention and recovers the local details at the end. The input image $\textbf{x} \in \mathbb{R}^{H\times W\times 3}$ is first evenly divided into $N =\frac{H}{16} \times \frac{W}{16}$ patches. Then the consecutive $3 \times 3 \times 3$ upsampling-convolution layers is used to obtain $\textbf{z}^1\in \mathbb{R}^{\frac{H}{16} \times \frac{W}{16}\times D_1 }$, $\textbf{z}^2\in \mathbb{R}^{\frac{H}{8} \times \frac{W}{8}\times D_2 }$, and $\textbf{z}^3\in \mathbb{R}^{\frac{H}{4} \times \frac{W}{4}\times D_3 }$. We propose different models with transformer variants.
3. BiFusion Module
It fuses the encoded features of the same resolution extracted from CNN and Transformer branch. The fused feature $\textbf{b}^i, i = 1,2,3$ is obtained by the following operations:
$\hat{\textbf{z}}^i = \text{ChannelAttn}({\textbf{z}}^i)$
$\hat{\textbf{l}}^i = \text{SpatialAttn}({\textbf{l}}^i)$
$\hat{\textbf{c}}^i = \text{Conv}(\textbf{z}^i\textbf{W}_1^i\odot\textbf{l}^i\textbf{W}_2^i)$
$\hat{\textbf{b}}^i = \text{Residual}([\hat{\textbf{c}}^i,\hat{\textbf{z}}^i,\hat{\textbf{l}}^i])$
where $\textbf{W}_1^i \in \mathbb{R}^{D_i\times L_i}$, $\textbf{W}_2^i \in \mathbb{R}^{C_i\times L_i}$, $|\odot|$ is the Hadamard product and Conv is a $3\times3$ convolution layer. The channel attention is implemented according to the SE-Block proposed by to facilitate global information from the Transformer branch. [10] The spatial attention is adopted from CBAM block as spatial filters to enhance local details. [11]
4. Attention-gated skip-connection
It combines fused feature maps and generates the segmentation. [12] We obtain $\hat{\textbf{f}}^{i+1} = \text{Conv}([\text{Up}(\hat{\textbf{f}}^{i}), \text{AG}({\textbf{f}}^{i+1}, \text{Up}(\hat{\textbf{f}}^{i}))])$ and $\textbf{f}^{1}=\hat{\textbf{f}}^{1}$.
With this branch-in-parallel approach, firstly, global information can be captured and sensitivity could be preserved on low-level context without building deep nets; secondly, the fused representation would be powerful and compact with the BiFusion module by utilizing the different features of CNN and Transformer simultaneously.
Swin-Fuse
In order to solve the problem of fragmental edges of Swin-UNet, the Swin-Transformer is optimized with the BiFusion module. By funding the encoder of ResNet and encoder od Swin-Transformer together, low level details captured by the CNN branch and the global information captured by the Transformer branch can be combined together by the BiFusion module and then sent to the decode/ The hybrid model therefore can preserve the strength of high prediction accuracy of Swin-UNet and further improve the model's ability of restoring edges of organs with the help of the captured low-level details.
Swin-Fuse is trained with ADA< optimizer with learning rate set to 0.001 Beta1 and Beta 2 are set to 0.5 and 0.999 correspondingly. The loss of the output is the weighted sum of the cross entropy and DICE score with weights of 0.4 and 0.6. The cross entropy and DICE loss are calculated based on the intermediate result and final result, consistent with the method introduced by the TransFuse.
UT-Fuse
In practice, all transformer-based models such as Trans-UNet, Swin-UNet, UTNet and hybrid models like TransFuse and Swin-Fuse failed to segment left adrenal gland and right adrenal gland. After analyzing the characteristics of these two organs, it is found that they are small and direction-dependent organs, that they are located almost in the same slice of the 3D CT scan, and only differentiating in the direction (left and right oriented) and the relative location in the 2D image. In order to successfully segment these two organs, the model's ability of classifying small organs based on the neighboring organ and also tell the direction of organs is needed to enhance.
Therefore, we proposed UT-Fuse, fusing ResNet and UTNet with the BiFusion module. The model is capable of capturing pixel-level details from the input image from both the transformer branch and the CNN branch. The transformer branch will also process the location information with global self-attention. Since both branches have an intermediate convolutional layer, neighboring organs have stronger connections and the model can detect direction dependent organs based on the relative location with other organs.
Training method and parameters of UT-Fuse is consistent with that of Swin-Fuse.
Implementation
Synapse multi-organ image segmentation dataset acts as the data in this project, that was originally used for the MICCAI 2015 Multi-Atlas Abdomen Labeling Challenge. [13] 50 abdomen CT scans are included in the dataset. To align with previous works, 30 out of 50 scans are used for this project. These CT scans range from $512 \times 512 \times 85$ to $512 \times 512 \times 198$ in pixels with resolution varying from $0.54 \times 0.54$ $mm^2$ to $0.98 \times 0.98$ $mm^2$. 13 different organs, including spleen, stomach, liver, etc. are labeled pixel by pixel.
3D CT scan is cut into $512 \times 512$ pixel 2D images, and splited 12 out of 30 scans as testing images, which is consistent to preprocessing techniques mentioned in SwinUNet and TranUNet. Models such as TranUNet and SwinUNet use pretrained weight on large image dataset while novel architectural models like UTNet use a unique down-sampling method that requires specific image dimension. Therefore, the image is cropped to [224,224] or [256, 256] in dimension corresponding to the model requirements. In order to improve the robustness of the model, data augmentation methods such as random flip and random rotation have been applied.
Evaluation
Though many transformer-based models reported better performance compared to traditional Res-UNet, only models of comparable sizes are used for evaluation. For example, Res-UNet based on ResNet-34 which has 63.5 million parameters and TransUNet based on vit-base-patch 16-224 which has 86 million parameters are used as reference models. The implemented hybrid model uses the pre-trained weight of ResNet-34 and Tiny Swin-Transformer (28 million parameters).
For medical images segmentation, there are two types of errors related to segmentation accuracy, namely the delineation of the boundary (contour) and the size (volume of the segmented object). Two metrics for these two types of errors are selected based on literature review. [14]
Dice Coefficient, also called the overlap index, is the most used metric in validating medical image segmentation, which is defined as
$DICE = \frac{2TP}{2TP + FP + FN}$
Hausdorff Distance(HD) measures the distance between crisp volumes. Given two point sets $A$ and $B$, HD is defined by
$HD(A, B) = \max(h(A, B), h(B,A))$
where $h(A, B)$ is called the directed Hausdorff distance and given by
$h(A, B) = \max_{a\in A}\min_{b\in B} ||a-b||$
where $||\cdot||$ is a norm(normally Euclidean distance). Note that HD is sensitive to outliers and the $q$-th quantile of distances may be used instead of the maximum, so that possible outliers are excluded.
Result Analysis
In this part, the average Dice Coefficient and average Hausdorff Distance(HD) on 13 organs will be reported. The organs included spleen, right kidney, left kidney, gallbladder, esophagus, liver, stomach, aorta, inferior vena cava, portal vein and splenic vein, pancreas, right adrenal gland, and left adrenal gland. They are splitted of 18 training cases (2212 axial slices) and 12 cases for validation. Table 1 and Table 2 showed the results comparison.
| DICE Score | ResUNet | TransUNet | SwinUNet | UTNet | TransFuse | Swin-Fuse | UT-Fuse |
|---|---|---|---|---|---|---|---|
| spleen | 81.43% | 83.43% | 87.49% | 86.46% | 86.07% | 89.45% | 87.10% |
| right kidney | 71.77% | 69.89% | 77.00% | 83.03% | 79.07% | 79.81% | 72.90% |
| left kidney | 80.12% | 74.83% | 81.15% | 86.12% | 79.08% | 86.64% | 82.11% |
| gallbladder | 61.17% | 45.57% | 60.04% | 70.97% | 26.68% | 63.96% | 58.67% |
| esophagus | 71.53% | 54.54% | 67.64% | 73.76% | 69.90% | 66.48% | 66.12% |
| liver | 93.71% | 91.51% | 93.42% | 95.42% | 93.79% | 93.21% | 93.92% |
| stomach | 73.92% | 72.43% | 72.46% | 75.50% | 71.45% | 73.36% | 80.70% |
| aorta | 87.39% | 70.97% | 83.18% | 89.99% | 85.94% | 86.34% | 87.34% |
| inferior vena cava | 74.47% | 59.69% | 70.57% | 81.33% | 76.52% | 77.59% | 69.65% |
| portal vein and splenic vein | 66.32% | 39.60% | 54.13% | 66.69% | 55.67% | 57.43% | 62.08% |
| pancreas | 61.22% | 45.00% | 54.81% | 62.34% | 62.30% | 56.49% | 61.72% |
| right adrenal gland | 59.26% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 45.91% |
| left adrenal gland | 58.21% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 50.33% |
| Mean | 72.35% | 54.42% | 61.68% | 67.05% | 60.50% | 63.90% | 70.66% |
Table 1: Mean Dice by organs
| DICE Score | ResUNet | TransUNet | SwinUNet | UTNet | TransFuse | Swin-Fuse | UT-Fuse |
|---|---|---|---|---|---|---|---|
| spleen | 68.37 | 31.07 | 29.63 | 35.77 | 34.35 | 33.60 | 8.78 |
| right kidney | 74.55 | 62.91 | 31.54 | 54.66 | 41.48 | 19.52 | 39.99 |
| left kidney | 39.32 | 40.32 | 47.11 | 35.62 | 22.19 | 16.18 | 39.22 |
| gallbladder | 24.36 | 38.71 | 29.26 | 10.95 | 10.21 | 10.85 | 14.52 |
| esophagus | 6.52 | 7.94 | 5.93 | 5.14 | 4.61 | 8.78 | 6.11 |
| liver | 26.54 | 25.21 | 21.05 | 12.77 | 14.70 | 27.26 | 20.18 |
| stomach | 23.10 | 17.44 | 18.28 | 17.64 | 25.85 | 16.32 | 11.90 |
| aorta | 15.49 | 11.59 | 10.69 | 4.27 | 10.96 | 11.90 | 6.35 |
| inferior vena cava | 8.30 | 18.33 | 12.89 | 5.43 | 9.28 | 6.16 | 8.86 |
| portal vein and splenic vein | 15.50 | 32.08 | 29.13 | 17.45 | 16.72 | 29.02 | 23.51 |
| pancreas | 10.63 | 20.35 | 15.48 | 12.62 | 12.45 | 20.80 | 11.13 |
| right adrenal gland | 5.31 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 7.55 |
| left adrenal gland | 5.94 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 6.40 |
| Mean | 24.92 | 23.54 | 22.82 | 19.30 | 18.44 | 18.22 | 15.73 |
Table 2: Mean hd95 by organs
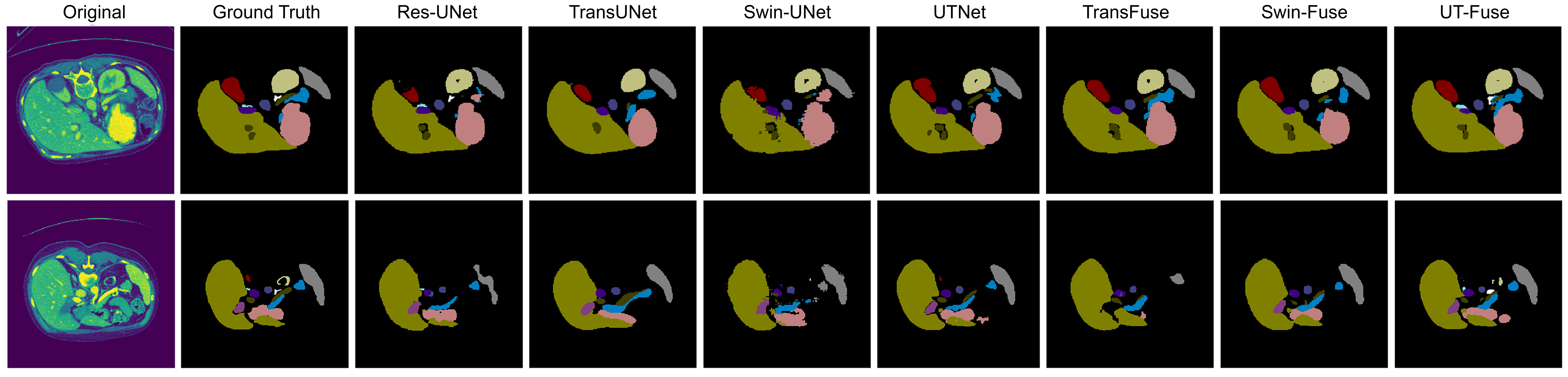
Fig 2. - Segmentation result of Synapse dataset.
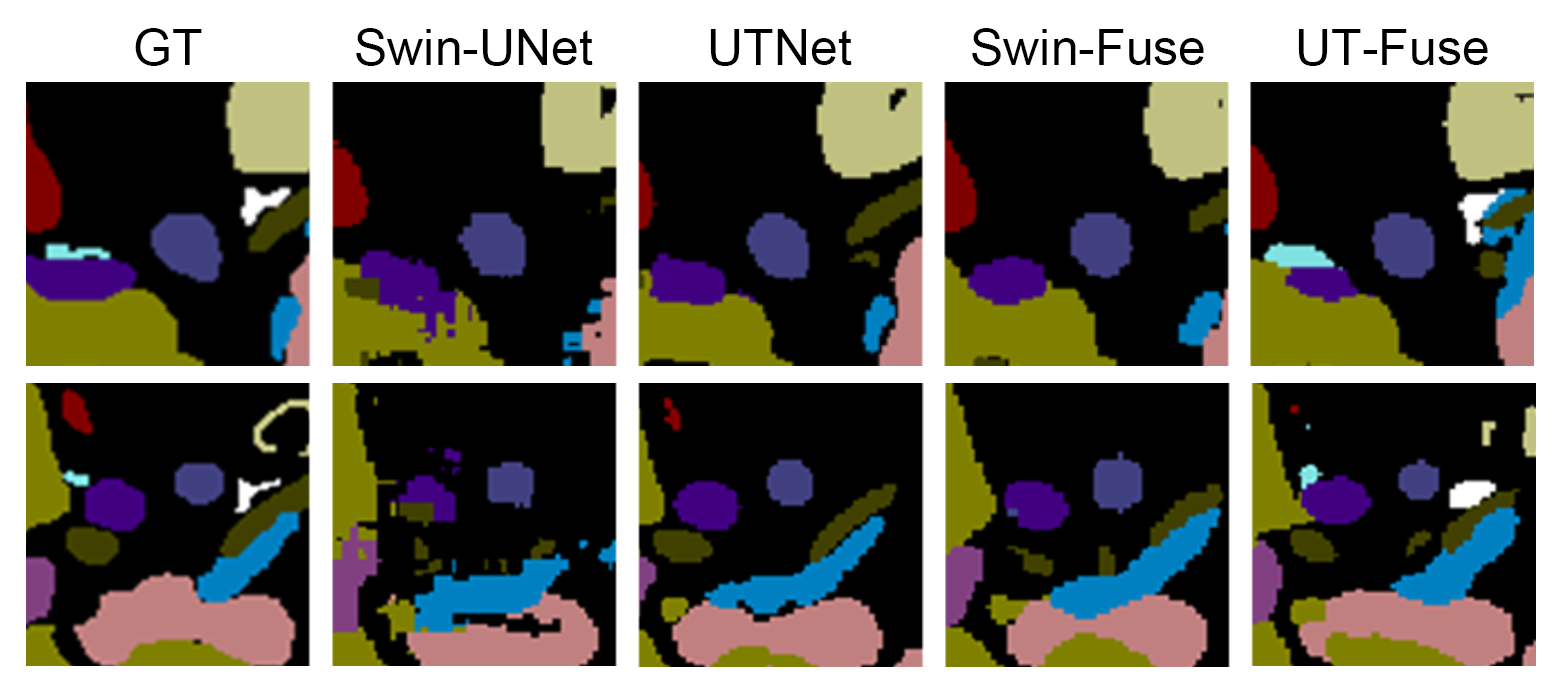
Fig 3. - Local segmentation result for small objects
Swin-Fuse
With the help of BiFusion module and CNN-branch, Swin-Fuse has better performance in capturing the local interactions with neighbouring pixels and therefore it can restore the edge of detected objects better. According to our result, the classified organs from Swin-Fuse could be properly shaped, while edged from Swin-UNet is fragmental, as shown in Fig.2. Comparing mean DICE score and mean HD95 of Swin-Fuse(63.90%, 18.22) and TransUNet(54.42%, 23.54), Swin-UNet has a better performance.
UT-Fuse
UT-Fuse which is fused by hybrid UTNet and ResNet-34 has a better DICE score compared to other Transformer-based models. With a margin of 3.61\% higher than UTNet (67.05\%) and 16.24\% higher than the original TransUNet. The HD95 of UT-Fuse is also better than other models. The mean DICE score of UT-Fuse is 15.73, beating the second place, Swin-Fuse, by 18.22. UT-Net can also classify and segment small and direction dependent objects such as right and left adrenal gland which all other Transformer-based models failed to segment, with DICE score of 45.91\% and 50.33\%, and HD95 of 7.55 and 6.40. Though UT-Fuse has a lower DICE score than Res-UNet (70.66\% compared to 72.35\%), the HD95 of UT-Fuse (15.73) is much better than that of Res-UNet (24.92).
The performance of UT-Fuse is generally better than almost all other models, however, it is inferior to UTNet in several classes. An interesting observation is that both Res-UNet and UTNet have higher DICE scores for gallbladder than UT-Fuse even though UT-Fuse is composed of these two models. This problem is possibly caused by the unclear boundary of gallbladder with other organs, when the BiFusion module fusing two separate models, information captured by lower layers are mixed up, which makes the model harder to separate Gallbladder and other organs.
Reference
[1] Sihang Zhou, Dong Nie, Ehsan Adeli, Jianping Yin, Jun Lian, and Dinggang Shen. 2019. High-resolution encoder-decoder networks for low-contrast medical image segmentation. IEEE Transactions on Image Processing, 29:461-475.
[2] Olaf Ronneberger, Philipp Fischer, and Thomas Brox. 2015. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention, pages 234-241. Springer.
[3] Zhengxin Zhang, Qingjie Liu, and Yunhong Wang. 2018. Road extraction by deep residual u-net. IEEE Geoscience and Remote Sensing Letters, 15(5):749-753.
[4] Kaiming He, Xiangyu Zhang, Shaoqing Ren, and Jian Sun. 2016. Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 770-778.
[5] Jieneng Chen, Yongyi Lu, Qihang Yu, Xiangde Luo, Ehsan Adeli, Yan Wang, Le Lu, Alan L Yuille, and Yuyin Zhou. 2021. Transunet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306.
[6] Hu Cao, Yueyue Wang, Joy Chen, Dongsheng Jiang, Xiaopeng Zhang, Qi Tian, and Manning Wang. 2021. Swin-unet: Unet-like pure transformer for medical image segmentation.
[7] Yunhe Gao, Mu Zhou, and Dimitris N Metaxas. 2021. Utnet: a hybrid transformer architecture for medical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pages 61-71. Springer.
[8] Ali Hatamizadeh, Yucheng Tang, Vishwesh Nath, Dong Yang, Andriy Myronenko, Bennett Landman, Holger Roth, and Daguang Xu. 2021. Unetr: Transformers for 3d medical image segmentation.
[9] Yundong Zhang, Huiye Liu, and Qiang Hu. 2021. Transfuse: Fusing transformers and cnns for medical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pages 14-24. Springer.
[10] Jie Hu, Li Shen, and Gang Sun. 2018. Squeeze-andexcitation networks. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 7132-7141.
[11] Sanghyun Woo, Jongchan Park, Joon-Young Lee, and In So Kweon. 2018. Cbam: Convolutional block attention module. In Proceedings of the European conference on computer vision (ECCV), pages 3-19.
[12] Jo Schlemper, Ozan Oktay, Michiel Schaap, Mattias Heinrich, Bernhard Kainz, Ben Glocker, and Daniel Rueckert. 2019. Attention gated networks: Learning to leverage salient regions in medical images. Medical image analysis, 53:197-207.
[13] [email protected] Sage Bionetworks. Sage bionetworks.
[14] Abdel Aziz Taha and Allan Hanbury. 2015. Metrics for evaluating 3d medical image segmentation: analysis, selection, and tool. BMC medical imaging, 15(1):1-28.
